Hauptinhalt
Topinformationen
Molekulare Mechanismen bakterieller Pathogenität

Prof. Dr. Michael Hensel, Dipl.-Biol.
Tel.: +49 541 969-3940
Fax: +49 541 969-3942
Sprechzeiten: n.V.
Raum: 36/121
e-Mail
2020
Elpers, L., & Hensel, M. (2020). Expression and functional characterization of various chaperon-usher fimbriae, Curli fimbriae, and Type 4 Pili of Enterohemorrhagic Escherichia coli O157:H7 Sakai. Front Microbiol, 11. doi:10.3389/fmicb.2020.00378
Elpers, L., Kretzschmar, J., Nuccio, S. P., Baumler, A. J., & Hensel, M. (2020). Factors Required for Adhesion of Salmonella enterica Serovar Typhimurium to Corn Salad (Valerianella locusta). Appl Environ Microbiol, 86(8). doi:10.1128/AEM.02757-19
Galeev, A., Suwandi, A., Bakker, H., Oktiviyari, A., Routier, F. H., Krone, L., . . . Grassl, G. A. (2020). Proteoglycan-dependent endo-lysosomal fusion affects intracellular survival of Salmonella Typhimurium in epithelial cells. Frontiers in Immunology, accepted.
Göser, V., & Hensel, M. (2020). Self-labeling enzyme tags for translocation analyses of Salmonella effector proteins. Methods Mol Biol, in press.
Göser, V., Kehl, A., Roder, J., & Hensel, M. (2020). Role of the ESCRT-III complex in controlling integrity of the Salmonella-containing vacuole. Cell Microbiol, in press, e13176. doi:10.1111/cmi.13176
Göser, V., Kommnick, C., Liss, V., & Hensel, M. (2019). Self-Labeling Enzyme Tags for Analyses of Translocation of Type III Secretion System Effector Proteins. mbio, 10(3). doi:10.1128/mBio.00769-19
Kehl, A., Noster, J., & Hensel., M. (2020). Eat in or take out? Metabolism of intracellular Salmonella enterica. Trends Microbiol, in press.
Kirchweger, P., Weiler, S., Egerer-Sieber, C., Blasl, A. T., Hoffmann, S., Schmidt, C., . . . Muller, Y. A. (2019). Structural and functional characterization of SiiA, an auxiliary protein from the SPI4-encoded type 1 secretion system from Salmonella enterica. Mol Microbiol, 112(5), 1403-1422. doi:10.1111/mmi.14368
Klingl, S., Kordes, S., Schmid, B., Gerlach, R. G., Hensel, M., & Muller, Y. A. (2020). Recombinant protein production and purification of SiiD, SiiE and SiiF - Components of the SPI4-encoded type I secretion system from Salmonella Typhimurium. Protein Expr Purif, accepted, 105632. doi:10.1016/j.pep.2020.105632
Kommnick, C., & Hensel, M. (2020). Correlative light and scanning electron microscopy for adhesion/invasion studies of Salmonella Typhimurium in polarized monolayer cells. Meth. Mol. Biol., in press.
2019
Kommnick, C., Lepper, A., & Hensel, M. (2019). Correlative light and scanning electron microscopy (CLSEM) for analysis of bacterial infection of polarized epithelial cells. Sci Rep, 9(1), 17079.
doi:10.1038/s41598-019-53085-6
Noster, J., Chao, T. C., Sander, N., Schulte, M., Reuter, T., Hansmeier, N., & Hensel, M. (2019). Proteomics of intracellular Salmonella enterica reveals roles of Salmonella pathogenicity island 2 in metabolism and antioxidant defense. PLoS Pathog, 15(4), e1007741. doi:10.1371/journal.ppat.1007741
Noster, J., Hansmeier, N., Persicke, M., Chao, T. C., Kurre, R., Popp, J., . . . Hensel, M. (2019). Blocks in Tricarboxylic Acid Cycle of Salmonella enterica Cause Global Perturbation of Carbon Storage, Motility, and Host-Pathogen Interaction. mSphere, 4(6). doi:10.1128/mSphere.00796-19
Noster, J., Persicke, M., Chao, T. C., Krone, L., Heppner, B., Hensel, M., & Hansmeier, N. (2019). Impact of ROS-Induced Damage of TCA Cycle Enzymes on Metabolism and Virulence of Salmonella enterica serovar Typhimurium. Front Microbiol, 10, 762. doi:10.3389/fmicb.2019.00762
Roder, J., & Hensel, M. (2020). Presence of SopE and mode of infection result in increased Salmonella-containing vacuole damage and cytosolic release during host cell infection by Salmonella enterica. Cell Microbiol, 22(5), e13155. doi:10.1111/cmi.13155
Schulte, M., Sterzenbach, T., Miskiewicz, K., Elpers, L., Hensel, M., & Hansmeier, N. (2019). A versatile remote control system for functional expression of bacterial virulence genes based on the tetA promoter. Int J Med Microbiol, 309(1), 54-65. doi:10.1016/j.ijmm.2018.11.001
Suwandi, A., Galeev, A., Riedel, R., Sharma, S., Seeger, K., Sterzenbach, T., . . . Grassl, G. A. (2019). Std fimbriae-fucose interaction increases Salmonella-induced intestinal inflammation and prolongs colonization. PLoS Pathog, 15(7), e1007915. doi:10.1371/journal.ppat.1007915
2018
Brink, T., Leiss, V., Siegert, P., Jehle, D., Ebner, J. K., Schwan, C., . . . Orth, J. H. C. (2018). Salmonella Typhimurium effector SseI inhibits chemotaxis and increases host cell survival by deamidation of heterotrimeric Gi proteins. PLoS Pathog, 14(8), e1007248. doi:10.1371/journal.ppat.1007248
Fulde, M., Sommer, F., Chassaing, B., van Vorst, K., Dupont, A., Hensel, M., . . . Hornef, M. W. (2018). Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature, 560(7719), 489-493. doi:10.1038/s41586-018-0395-5
Haas, A., Hensel, M., Luhrmann, A., Rudel, T., Saftig, P., & Schaible, U. E. (2018). Intracellular compartments of pathogens: Highways to hell or stairways to heaven? Int J Med Microbiol, 308(1), 1-2.
doi:10.1016/j.ijmm.2017.10.009
Namakchian, M., Kassler, K., Sticht, H., Hensel, M., & Deiwick, J. (2018). Structure-based functional analysis of effector protein SifA in living cells reveals motifs important for Salmonella intracellular proliferation. Int J Med Microbiol, 308(1), 84-96. doi:10.1016/j.ijmm.2017.09.004
Sprenger, M., Kasper, L., Hensel, M., & Hube, B. (2018). Metabolic adaptation of intracellular bacteria and fungi to macrophages. Int J Med Microbiol, 308(1), 215-227. doi:10.1016/j.ijmm.2017.11.001
Zhang, K., Riba, A., Nietschke, M., Torow, N., Repnik, U., Putz, A., . . . Hornef, M. (2018). Minimal SPI1-T3SS effector requirement for Salmonella enterocyte invasion and intracellular proliferation in vivo. PLoS Pathog, 14(3), e1006925. doi:10.1371/journal.ppat.1006925
2017
Aviv, G., Elpers, L., Mikhlin, S., Cohen, H., Vitman Zilber, S., Grassl, G. A., . . . Gal-Mor, O. (2017). The plasmid-encoded Ipf and Klf fimbriae display different expression and varying roles in the virulence of Salmonella enterica serovar Infantis in mouse vs. avian hosts. PLoS Pathog, 13(8), e1006559.
doi:10.1371/journal.ppat.1006559
Fabiani, F. D., Renault, T. T., Peters, B., Dietsche, T., Galvez, E. J. C., Guse, A., . . . Erhardt, M. (2017). A flagellum-specific chaperone facilitates assembly of the core type III export apparatus of the bacterial flagellum. PLoS Biol, 15(8), e2002267. doi:10.1371/journal.pbio.2002267
Hansmeier, N., Miskiewicz, K., Elpers, L., Liss, V., Hensel, M., & Sterzenbach, T. (2017). Functional expression of the entire adhesiome of Salmonella enterica serotype Typhimurium. Sci Rep, 7(1), 10326.
doi:10.1038/s41598-017-10598-2
Liss, V., Swart, A. L., Kehl, A., Hermanns, N., Zhang, Y., Chikkaballi, D., . . . Hensel, M. (2017). Salmonella enterica remodels the host cell endosomal system for efficient intravacuolar nutrition. Cell Host Microbe, 21(3), 390-402. doi:10.1016/j.chom.2017.02.005
Peters, B., Stein, J., Klingl, S., Sander, N., Sandmann, A., Taccardi, N., . . . Hensel, M. (2017). Structural and functional dissection reveals distinct roles of Ca2+-binding sites in the giant adhesin SiiE of Salmonella enterica. PLoS Pathog, 13(5), e1006418. doi:10.1371/journal.ppat.1006418
2016
Barlag, B., Beutel, O., Janning, D., Czarniak, F., Richter, C. P., Kommnick, C., . . . Hensel, M. (2016). Single molecule super-resolution imaging of proteins in living Salmonella enterica using self-labelling enzymes. Sci Rep, 6, 31601. doi:10.1038/srep31601
Klionsky, D. J., Abdelmohsen, K., Abe, A., Abedin, M. J., Abeliovich, H., Acevedo Arozena, A., . . . Zughaier, S. M. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy, 12(1), 1-222. doi:10.1080/15548627.2015.1100356
Ortiz de Orue Lucana, D., Hickey, N., Hensel, M., Klare, J. P., Geremia, S., Tiufiakova, T., & Torda, A. E. (2016). The Crystal Structure of the C-Terminal Domain of the Salmonella enterica PduO Protein: An Old Fold with a New Heme-Binding Mode. Front Microbiol, 7, 1010. doi:10.3389/fmicb.2016.01010
Schulte, M., & Hensel, M. (2016). Models of intestinal infection by Salmonella enterica: introduction of a new neonate mouse model. F1000Res, 5. doi:10.12688/f1000research.8468.1
2015
Barlag, B., & Hensel, M. (2015). The giant adhesin SiiE of Salmonella enterica. Molecules, 20(1), 1134-1150. doi:10.3390/molecules20011134
Czarniak, F., & Hensel, M. (2015). Red-mediated recombineering of Salmonella enterica genomes. Methods Mol Biol, 1225, 63-79. doi:10.1007/978-1-4939-1625-2_4
Frahm, M., Felgner, S., Kocijancic, D., Rohde, M., Hensel, M., Curtiss, R., 3rd, . . . Weiss, S. (2015). Efficiency of conditionally attenuated Salmonella enterica serovar Typhimurium in bacterium-mediated tumor therapy. mbio, 6(2). doi:10.1128/mBio.00254-15
Hegazy, W. A., Xu, X., Metelitsa, L., & Hensel, M. (2015). Correction for Hegazy et al., Evaluation of Salmonella enterica Type III Secretion System Effector Proteins as Carriers for Heterologous Vaccine Antigens. Infect Immun, 83(3), 1225. doi:10.1128/IAI.03010-14
Herweg, J. A., Hansmeier, N., Otto, A., Geffken, A. C., Subbarayal, P., Prusty, B. K., . . . Hilbi, H. (2015). Purification and proteomics of pathogen-modified vacuoles and membranes. Front Cell Infect Microbiol, 5, 48. doi:10.3389/fcimb.2015.00048
Jennewein, J., Matuszak, J., Walter, S., Felmy, B., Gendera, K., Schatz, V., . . . Jantsch, J. (2015). Low-oxygen tensions found in Salmonella-infected gut tissue boost Salmonella replication in macrophages by impairing antimicrobial activity and augmenting Salmonella virulence. Cell Microbiol, 17(12), 1833-1847.
doi:10.1111/cmi.12476
Kehl, A., & Hensel, M. (2015). Live cell imaging of intracellular Salmonella enterica. Methods Mol Biol, 1225, 199-225. doi:10.1007/978-1-4939-1625-2_13
Liss, V., Barlag, B., Nietschke, M., & Hensel, M. (2015). Self-labelling enzymes as universal tags for fluorescence microscopy, super-resolution microscopy and electron microscopy. Sci Rep, 5, 17740.
doi:10.1038/srep17740
Liss, V., & Hensel, M. (2015). Take the tube: remodelling of the endosomal system by intracellular Salmonella enterica. Cell Microbiol, 17(5), 639-647. doi:10.1111/cmi.12441
Popp, J., Noster, J., Busch, K., Kehl, A., Zur Hellen, G., & Hensel, M. (2015). Role of Host Cell-Derived Amino Acids in Nutrition of Intracellular Salmonella enterica. Infect Immun, 83(12), 4466-4475.
doi:10.1128/IAI.00624-15
Vorwerk, S., Krieger, V., Deiwick, J., Hensel, M., & Hansmeier, N. (2015). Proteomes of host cell membranes modified by intracellular activities of Salmonella enterica. Mol Cell Proteomics, 14(1), 81-92.
doi:10.1074/mcp.M114.041145
Wille, T., Barlag, B., Jakovljevic, V., Hensel, M., Sourjik, V., & Gerlach, R. G. (2015). A gateway-based system for fast evaluation of protein-protein interactions in bacteria. PLoS One, 10(4), e0123646.
doi:10.1371/journal.pone.0123646
Zhang, Y., Krieger, V., & Hensel, M. (2015). Application of fluorescent nanoparticles to study remodeling of the endo-lysosomal system by intracellular bacteria. J Vis Exp, 95(95), e52058. doi:10.3791/52058
2014
Ahmed, Z., Zeeshan, S., Huber, C., Hensel, M., Schomburg, D., Munch, R., . . . Dandekar, T. (2014). 'Isotopo' a database application for facile analysis and management of mass isotopomer data. Database (Oxford), 2014. doi:10.1093/database/bau077
Bohles, N., Busch, K., & Hensel, M. (2014). Vaccines against human diarrheal pathogens: Current status and perspectives. Hum Vaccin Immunother, 10(6), 1522-1535. doi:10.4161/hv.29241
Dandekar, T., Fieselmann, A., Fischer, E., Popp, J., Hensel, M., & Noster, J. (2014). Salmonella-how a metabolic generalist adopts an intracellular lifestyle during infection. Front Cell Infect Microbiol, 4, 191.
doi:10.3389/fcimb.2014.00191
Grin, I., Hartmann, M. D., Sauer, G., Hernandez Alvarez, B., Schutz, M., Wagner, S., . . . Linke, D. (2014). A trimeric lipoprotein assists in trimeric autotransporter biogenesis in enterobacteria. J Biol Chem, 289(11), 7388-7398. doi:10.1074/jbc.M113.513275
Husseiny, M. I., Rawson, J., Kaye, A., Nair, I., Todorov, I., Hensel, M., . . . Ferreri, K. (2014). An oral vaccine for type 1 diabetes based on live attenuated Salmonella. Vaccine, 32(20), 2300-2307.
doi:10.1016/j.vaccine.2014.02.070
Krieger, V., Liebl, D., Zhang, Y., Rajashekar, R., Chlanda, P., Giesker, K., . . . Hensel, M. (2014). Reorganization of the endosomal system in Salmonella-infected cells: the ultrastructure of Salmonella-induced tubular compartments. PLoS Pathog, 10(9), e1004374. doi:10.1371/journal.ppat.1004374
Lorkowski, M., Felipe-Lopez, A., Danzer, C. A., Hansmeier, N., & Hensel, M. (2014). Salmonella enterica invasion of polarized epithelial cells is a highly cooperative effort. Infect Immun, 82(6), 2657-2667.
doi:10.1128/IAI.00023-14
Rajashekar, R., Liebl, D., Chikkaballi, D., Liss, V., & Hensel, M. (2014). Live cell imaging reveals novel functions of Salmonella enterica SPI2-T3SS effector proteins in remodeling of the host cell endosomal system. PLoS One, 9(12), e115423. doi:10.1371/journal.pone.0115423
Wagner, C., Barlag, B., Gerlach, R. G., Deiwick, J., & Hensel, M. (2014). The Salmonella enterica giant adhesin SiiE binds to polarized epithelial cells in a lectin-like manner. Cell Microbiol, 16(6), 962-975.
doi:10.1111/cmi.12253
Wille, T., Wagner, C., Mittelstadt, W., Blank, K., Sommer, E., Malengo, G., . . . Gerlach, R. G. (2014). SiiA and SiiB are novel type I secretion system subunits controlling SPI4-mediated adhesion of Salmonella enterica. Cell Microbiol, 16(2), 161-178. doi:10.1111/cmi.12222
Xu, X., Hegazy, W. A., Guo, L., Gao, X., Courtney, A. N., Kurbanov, S., . . . Metelitsa, L. S. (2014). Effective cancer vaccine platform based on attenuated salmonella and a type III secretion system. Cancer Res, 74(21), 6260-6270. doi:10.1158/0008-5472.CAN-14-1169
Zhang, K., Dupont, A., Torow, N., Gohde, F., Leschner, S., Lienenklaus, S., . . . Hornef, M. W. (2014). Age-dependent enterocyte invasion and microcolony formation by Salmonella. PLoS Pathog, 10(9), e1004385.
doi:10.1371/journal.ppat.1004385
2013
Ahmed, Z., Zeeshan, S., Huber, C., Hensel, M., Schomburg, D., Munch, R., . . . Dandekar, T. (2013). Software LS-MIDA for efficient mass isotopomer distribution analysis in metabolic modelling. BMC Bioinformatics, 14, 218. doi:10.1186/1471-2105-14-218
Griessl, M. H., Schmid, B., Kassler, K., Braunsmann, C., Ritter, R., Barlag, B., . . . Muller, Y. A. (2013). Structural insight into the giant Ca2+-binding adhesin SiiE: implications for the adhesion of Salmonella enterica to polarized epithelial cells. Structure 21(5), 741-752. doi:10.1016/j.str.2013.02.020
Hensel, M., Klingauf, J., & Piehler, J. (2013). Imaging the invisible: resolving cellular microcompartments by superresolution microscopy techniques. Biol Chem, 394(9), 1097-1113. doi:10.1515/hsz-2012-0324
Zhang, Y., & Hensel, M. (2013). Evaluation of nanoparticles as endocytic tracers in cellular microbiology. Nanoscale, 5(19), 9296-9309. doi:10.1039/c3nr01550e
2012
Dandekar, T., Astrid, F., Jasmin, P., & Hensel, M. (2012). Salmonella enterica: a surprisingly well-adapted intracellular lifestyle. Front Microbiol, 3, 164. doi:10.3389/fmicb.2012.00164
Hegazy, W. A., & Hensel, M. (2012). Salmonella enterica as a vaccine carrier. Future Microbiol, 7(1), 111-127. doi:10.2217/fmb.11.144
Hegazy, W. A., Xu, X., Metelitsa, L., & Hensel, M. (2012). Evaluation of Salmonella enterica type III secretion system effector proteins as carriers for heterologous vaccine antigens. Infect Immun, 80(3), 1193-1202. doi:10.1128/IAI.06056-11
Holzer, S. U., & Hensel, M. (2012). Divergent roles of Salmonella pathogenicity island 2 and metabolic traits during interaction of S. enterica serovar typhimurium with host cells. PLoS One, 7(3), e33220.
doi:10.1371/journal.pone.0033220
Müller, P., Chikkaballi, D., & Hensel, M. (2012). Functional dissection of SseF, a membrane-integral effector protein of intracellular Salmonella enterica. PLoS One, 7(4), e35004. doi:10.1371/journal.pone.0035004
Swart, A. L., & Hensel, M. (2012). Interactions of Salmonella enterica with dendritic cells. Virulence, 3(7), 660-667. doi:10.4161/viru.22761
Ustun, S., Müller, P., Palmisano, R., Hensel, M., & Bornke, F. (2012). SseF, a type III effector protein from the mammalian pathogen Salmonella enterica, requires resistance-gene-mediated signalling to activate cell death in the model plant Nicotiana benthamiana. New Phytol, 194(4), 1046-1060.
doi:10.1111/j.1469-8137.2012.04124.x
Wiese, M., Gerlach, R. G., Popp, I., Matuszak, J., Mahapatro, M., Castiglione, K., . . . Jantsch, J. (2012). Hypoxia-mediated impairment of the mitochondrial respiratory chain inhibits the bactericidal activity of macrophages. Infect Immun, 80(4), 1455-1466. doi:10.1128/IAI.05972-11
2011
Blank, K., Hensel, M., & Gerlach, R. G. (2011). Rapid and highly efficient method for scarless mutagenesis within the Salmonella enterica chromosome. PLoS One, 6(1), e15763. doi:10.1371/journal.pone.0015763
Creuzburg, K., Heeren, S., Lis, C. M., Kranz, M., Hensel, M., & Schmidt, H. (2011). Genetic background and mobility of variants of the gene nleA in attaching and effacing Escherichia coli. Appl Environ Microbiol, 77(24), 8705-8713. doi:10.1128/AEM.06492-11
Crull, K., Rohde, M., Westphal, K., Loessner, H., Wolf, K., Felipe-Lopez, A., . . . Weiss, S. (2011). Biofilm formation by Salmonella enterica serovar Typhimurium colonizing solid tumours. Cell Microbiol, 13(8), 1223-1233. doi:10.1111/j.1462-5822.2011.01612.x
Held, C., Palmisano, R., Haberle, L., Hensel, M., & Wittenberg, T. (2011). Comparison of parameter-adapted segmentation methods for fluorescence micrographs. Cytometry A, 79(11), 933-945.
doi:10.1002/cyto.a.21122
Jantsch, J., Chikkaballi, D., & Hensel, M. (2011). Cellular aspects of immunity to intracellular Salmonella enterica. Immunol Rev, 240(1), 185-195. doi:10.1111/j.1600-065X.2010.00981.x
Jantsch, J., Wiese, M., Schodel, J., Castiglione, K., Glasner, J., Kolbe, S., . . . Willam, C. (2011). Toll-like receptor activation and hypoxia use distinct signaling pathways to stabilize hypoxia-inducible factor 1alpha (HIF1A) and result in differential HIF1A-dependent gene expression. J Leukoc Biol, 90(3), 551-562.
doi:10.1189/jlb.1210683
Manuel, E. R., Blache, C. A., Paquette, R., Kaltcheva, T. I., Ishizaki, H., Ellenhorn, J. D., . . . Diamond, D. J. (2011). Enhancement of cancer vaccine therapy by systemic delivery of a tumor-targeting Salmonella-based STAT3 shRNA suppresses the growth of established melanoma tumors. Cancer Res, 71(12), 4183-4191. doi:10.1158/0008-5472.CAN-10-4676
Rajashekar, R., & Hensel, M. (2011). Dynamic modification of microtubule-dependent transport by effector proteins of intracellular Salmonella enterica. Eur J Cell Biol, 90(11), 897-902.
doi:10.1016/j.ejcb.2011.05.008
Schwidder, M., Hensel, M., & Schmidt, H. (2011). Regulation of nleA in Shiga toxin-producing Escherichia coli O84:H4 strain 4795/97. J Bacteriol, 193(4), 832-841. doi:10.1128/JB.00582-10
Sturm, K. U., Griessl, M. H., Wagner, C., Deiwick, J., Hensel, M., & Muller, Y. A. (2011). Crystallization and preliminary crystallographic analysis of an Ig-domain-encompassing fragment of the giant adhesion protein SiiE from Salmonella enterica. Acta Crystallogr Sect F Struct Biol Cryst Commun, 67(Pt 11), 1371-1374.
doi:10.1107/S1744309111032039
Wagner, C., & Hensel, M. (2011). Adhesive mechanisms of Salmonella enterica. Adv Exp Med Biol, 715, 17-34. doi:10.1007/978-94-007-0940-9_2
Wagner, C., Polke, M., Gerlach, R. G., Linke, D., Stierhof, Y. D., Schwarz, H., & Hensel, M. (2011). Functional dissection of SiiE, a giant non-fimbrial adhesin of Salmonella enterica. Cell Microbiol, 13(8), 1286-1301.
doi:10.1111/j.1462-5822.2011.01621.x
2010
Hölzer, S. U., & Hensel, M. (2010). Functional dissection of translocon proteins of the Salmonella pathogenicity island 2-encoded type III secretion system. BMC Microbiol, 10, 104. doi:10.1186/1471-2180-10-104
Ott, L., Holler, M., Gerlach, R. G., Hensel, M., Rheinlaender, J., Schaffer, T. E., & Burkovski, A. (2010). Corynebacterium diphtheriae invasion-associated protein (DIP1281) is involved in cell surface organization, adhesion and internalization in epithelial cells. BMC Microbiol, 10, 2. doi:10.1186/1471-2180-10-2
Ott, L., Holler, M., Rheinlaender, J., Schaffer, T. E., Hensel, M., & Burkovski, A. (2010). Strain-specific differences in pili formation and the interaction of Corynebacterium diphtheriae with host cells. BMC Microbiol, 10, 257. doi:10.1186/1471-2180-10-257
Wiese, M., Castiglione, K., Hensel, M., Schleicher, U., Bogdan, C., & Jantsch, J. (2010). Small interfering RNA (siRNA) delivery into murine bone marrow-derived macrophages by electroporation. J Immunol Methods, 353(1-2), 102-110. doi:10.1016/j.jim.2009.12.002
Xiong, G., Husseiny, M. I., Song, L., Erdreich-Epstein, A., Shackleford, G. M., Seeger, R. C., . . . Metelitsa, L. S. (2010). Novel cancer vaccine based on genes of Salmonella pathogenicity island 2. Int J Cancer, 126(11), 2622-2634. doi:10.1002/ijc.24957
Xu, X., & Hensel, M. (2010). Systematic analysis of the SsrAB virulon of Salmonella enterica. Infect Immun, 78(1), 49-58. doi:10.1128/IAI.00931-09
Xu, X., Husseiny, M. I., Goldwich, A., & Hensel, M. (2010). Efficacy of intracellular activated promoters for generation of Salmonella-based vaccines. Infect Immun, 78(11), 4828-4838. doi:10.1128/IAI.00298-10
2009
Duerr, C. U., Zenk, S. F., Chassin, C., Pott, J., Gutle, D., Hensel, M., & Hornef, M. W. (2009). O-antigen delays lipopolysaccharide recognition and impairs antibacterial host defense in murine intestinal epithelial cells. PLoS Pathog, 5(9), e1000567. doi:10.1371/journal.ppat.1000567
Gerlach, R. G., Jäckel, D., Hölzer, S. U., & Hensel, M. (2009). Rapid oligonucleotide-based recombineering of the chromosome of Salmonella enterica. Appl Environ Microbiol, 75(6), 1575-1580.
doi:10.1128/AEM.02509-08
Hölzer, S. U., Schlumberger, M. C., Jäckel, D., & Hensel, M. (2009). Effect of the O-antigen length of lipopolysaccharide on the functions of Type III secretion systems in Salmonella enterica. Infect Immun, 77(12), 5458-5470. doi:10.1128/IAI.00871-09
Husseiny, M. I., & Hensel, M. (2009). Evaluation of Salmonella live vaccines with chromosomal expression cassettes for translocated fusion proteins. Vaccine, 27(28), 3780-3787. doi:10.1016/j.vaccine.2009.03.053
Zenk, S. F., Jantsch, J., & Hensel, M. (2009). Role of Salmonella enterica lipopolysaccharide in activation of dendritic cell functions and bacterial containment. J Immunol, 183(4), 2697-2707.
doi:10.4049/jimmunol.0900937
Forschungsschwerpunkt
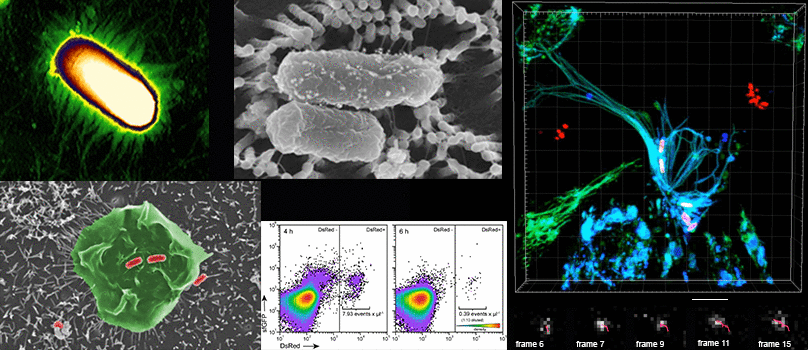
Die Arbeitsgruppe Hensel untersucht die molekularen Mechanismen bakterieller Pathogenität. Am Modellsystem des gastrointestinalen Erregers Salmonella enterica werden Mechanismen der Erkennung and Bindung von Wirtszellen, der Zellinvasion, und der intrazellulären Lebensweise in Wirtszellen untersucht.
Forschungsthemen
- Manipulation von Wirtszellen durch bakterielle Virulenzproteine
- Funktion von bakteriellen Proteinsekretionssystemen
Modellsysteme
- Salmonella enterica Serovar Typhimurium und isogene Mutanten
- Salmonella enterica Serovar Typhi und Papratyphi A
- Murine und humane Zelllinien und primäre Zellen
- Polarisierte Epithelzellen
- Intestinale Organoide
Methoden
- Lebendzellmikrokopie von Infektionsvorgängen
- Korrelative Licht- und Elektronenmikroskopie
Ausgewählte Publikationen
- Reuter T, Scharte F, Franzkoch R, Liss V, Hensel M. Single cell analyses reveal distinct adaptation of typhoidal and non-typhoidal Salmonella enterica serovars to intracellular lifestyle. PLoS pathogens. 2021;17(6):e1009319. DOI: 10.1371/journal.ppat.1009319 pdf
- Göser V, Kommnick C, Liss V, Hensel M. Self-labeling enzyme tags for analyses of translocation of type III secretion system effector proteins. mBio. 2019;10(3). DOI: 10.1128/mBio.00769-19. pdf
- Liss V, Swart AL, Kehl A, Hermanns N, Zhang Y, Chikkaballi D, et al. Salmonella enterica remodels the host cell endosomal system for efficient intravacuolar nutrition. Cell Host Microbe. 2017;21(3):390-402. DOI: 10.1016/j.chom.2017.02.005. pdf