Project Z1 | Florian Fröhlich & Joost Holthuis
Mass spectrometry, quantitative lipid analyses and lipid tools
This central project expands the consortium’s technical capabilities in analyzing lipid function by developing protocols for a rapid and efficient isolation of cellular organelles, establishing workflows for mass spectrometry-based analyses of organellar lipidomes and proteomes, and generating a toolbox of functionalized lipids for monitoring intracellular lipid flows, capturing lipid effector proteins, and manipulating subcellular lipid pools.
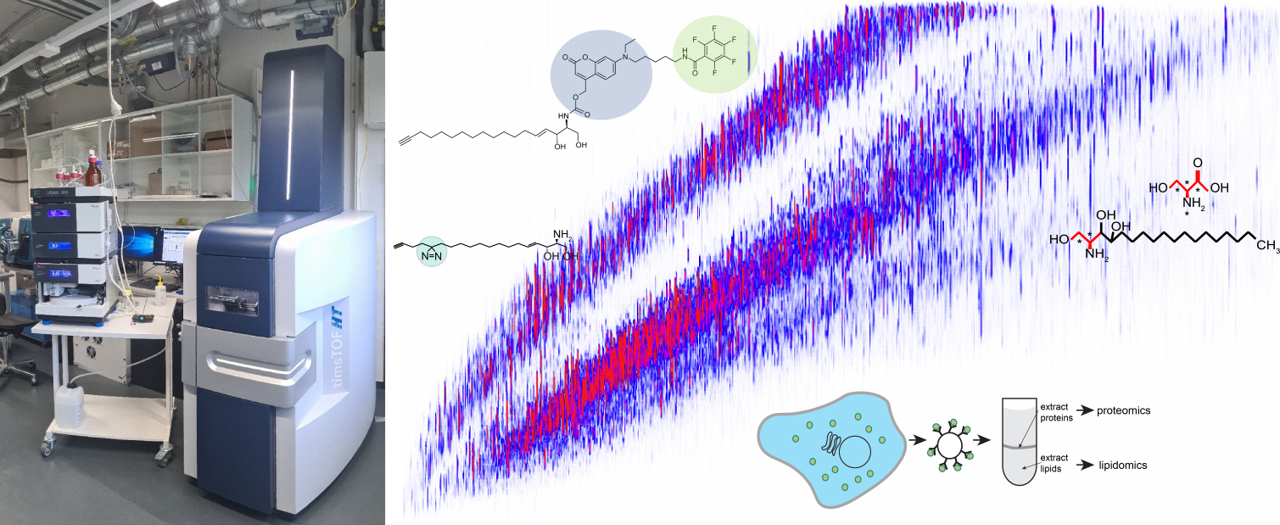
© Florian Fröhlich, Joost Holthuis & Stefan Walter
Project Summary
Prof. Dr. Florian Fröhlich
Osnabrück University
School of Biology/Chemistry
Bioanalytical Chemistry

Prof. Dr. Joost Holthuis
Osnabrück University
School of Biology/Chemistry
Molecular Cell Biology

Lipids are highly versatile biomolecules that form membrane barriers, participate in energy metabolism and serve as signalling molecules to control a wide array of cellular processes. However, assigning functions to individual lipid species is very challenging.
This complexity arises in part from the high interconnectivity of the lipid metabolic network, whereby changes in one lipid species can exert metabolic “ripple” effects with unexpected functional outcomes. Moreover, the lipid composition and trans-bilayer arrangement among organelles show striking variations, and there is compelling evidence that the collective properties of the bulk lipids play a profound part in defining organelle identity and function.
Even in one organelle, lipids can control different processes depending on whether they are localized in the inner or outer leaflet. The lipid metabolic network is coupled to an elaborate lipid transport system, collectively providing cells with an extremely rich repertoire for generating multiple signals and fine-tuning specific responses.
While lipids make a unique contribution to the plasticity of biomolecular networks, defining their subcellular distributions and biological roles also require unique experimental approaches, including methods to decipher and specifically alter organelle-specific lipidomes, map protein-lipid interactions and visualize lipid trafficking inside cells. To meet these challenges, the Z1 project will expand the consortium’s technical capabilities to analyze changes in protein and lipid abundance and distribution in cells.
Project-related Publications
Sokoya, T., Parolek, J., Foged, M.M., Danylchuk, D.I., Bozan, M., Sarkar, B., Hilderink, A., Philippi, M., Botto, L.D., Terhal, P.A., Mäkitie, O., Piehler, J., Kim, Y., Burd, C.G., Klymchenko, A.S., Maeda, K., Holthuis, J.C. (2022). Pathogenic variants of sphingomyelin synthase SMS2 disrupt lipid landscapes in the secretory pathway. Elife. 11: e79278.
Eising, S., Esch, B., Wälte, M., Vargas Duarte, P., Walter, S., Ungermann, C., Bohnert, M., Fröhlich, F. (2022). A lysosomal biogenesis map reveals the cargo spectrum of yeast vacuolar protein targeting pathways. J Cell Biol 221: e202107148.
Ponsford, A.H., Ryan, T.A., Raimondi, A., Cocucci, E., Wycislo, S.A., Fröhlich, F., Swan, L.E., Stagi, M. (2021). Live imaging of intra-lysosome pH in cell lines and primary neuronal culture using a novel genetically encoded biosensor. Autophagy 17, 1500–1518.
Morstein, J., Kol, M., Novak, A.J.E., Feng, S., Khayyo, S., Hinnah, K., Li-Purcell, N., Pan, G., Williams, B.M., Riezman, H., Atilla-Gokcumen, G.E., Holthuis, J.C., Trauner, D. (2021). Short Photoswitchable Ceramides Enable Optical Control of Apoptosis. ACS Chemical Biology 16, 452–456.
Dadsena, S., Bockelmann, S., Mina, J.G., Hassan, D., Korneev, S., Razzera, G., Jahn, H., Niekamp, P., Müller, D., Schneider, M., Tafesse, F.G., Marrink, S.J., Melo, M.N., Holthuis, J.C. (2019). Ceramides bind VDAC2 to trigger mitochondrial apoptosis. Nature Commun. 10, 1832.
Kol, M., Williams, B., Toombs-Ruane, H., Franquelim, H.G., Korneev, S., Schroeer, C., Schwille, P., Trauner, D†., Holthuis, J.C., Frank, J.A. (2019). Optical manipulation of sphingolipid biosynthesis using photoswitchable ceramides. Elife 8: e43230.
Eising, S., Thiele, L., Fröhlich, F. (2019). A systematic approach to identify recycling endocytic cargo depending on the garp complex. Elife 8: e42837.
Bockelmann, S., Mina, J.G.M., Korneev, S., Hassan, D.G., Müller, D., Hilderink, A., Vlieg, H.C., Raijmakers, R., Heck, A.J.R., Haberkant, P., Holthuis, J.C. (2018). A search for ceramide binding proteins using bifunctional lipid analogs yields CERT-related protein StarD7. J Lipid Res 59, 515–530.
Fröhlich, F., Olson, D.K., Christiano, R., Farese, R. v., Walther, T.C. (2016). Proteomic and phosphoproteomic analyses of yeast reveal the global cellular response to sphingolipid depletion. Proteomics 16, 2759–2763.
Guarani, V., McNeill, E.M., Paulo, J.A., Huttlin, E.L., Fröhlich, F., Gygi, S.P., Vactor, D. van, Wade Harper, J. (2015). QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. Elife 4: e06265.
Fröhlich, F., Christiano, R., Walther, T.C. (2013). Native SILAC: Metabolic labeling of proteins in prototroph microorganisms based on lysine synthesis regulation. Mol Cell Proteomics 12, 1995–2005.