Project P3 | Maria Bohnert
Plasticity of cellular lipid handling machineries in metabolic adaptation
We use microscopy-based screens to identify principles controlling the functional plasticity of lipid metabolism and inter-organelle lipid exchange, with a primary focus on sphingolipids, sterols, and storage lipids.
© Maria Bohnert
Project Summary
Prof. Dr. Maria Bohnert
University of Münster
Institute of Cell Dynamics and Imaging
Research Group Organelle Communication

Internal membranes divide eukaryotic cells into organelles, which allow for division of labor in cellular functions. On the flip side, these membranes also form obstacles to the flow of information and material through the cell.
How highly compartmentalized cells can respond to environmental cues in a precise, robust and coordinated manner is still poorly understood. Previous observations across various biological systems suggest that proteins involved in the handling of lipids, such as lipid metabolism enzymes and lipid transfer proteins, often change their localization and function in response to metabolic cues.
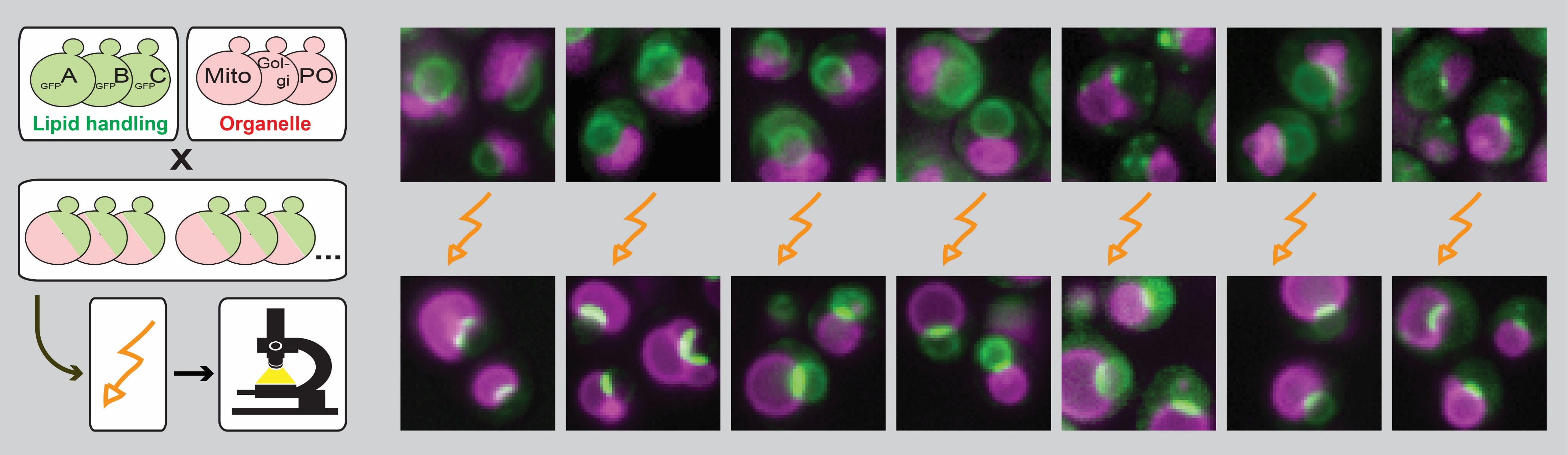
This adaptive behavior, which we here refer to as plasticity of lipid handling machineries, might be important for orchestrating coordinated responses across different organelles. We have developed an experimental platform that allows for systematic microscopic analysis of lipid handling proteins in response to metabolic cues and stress conditions in Saccharomyces cerevisiae.
In a pilot screen, we have found that the diacylglycerol acyltransferase Dga1, the sterol synthesis regulators Nsg1 and Nsg2, and the ceramide synthase subunits Lag1, Lac1, and Lip1 undergo dramatic relocations upon glucose starvation – from randomly dispersed to distinct foci on (putative) organelle contact sites. In this project, we will set out to broadly analyze the plasticity of lipid handling factors.
Project-related publications
Eising, S., Esch, B., Wälte, M., Vargas Duarte, P., Walter, S., Ungermann, C., Bohnert, M., Fröhlich, F. (2022). A lysosomal biogenesis map reveals the cargo spectrum of yeast vacuolar protein targeting pathways. J Cell Biol 221, e202107148.
Rampelt, H., Wollweber, F., Lecheva, M., de Boer, R., Perschil, I., Steidle, L., Becker, T., Bohnert, M., van der Laan, M., Pfanner, N. (2022). Dual role of Mic10 in mitochondrial cristae organization and ATP synthase-linked metabolic adaptation and respiratory growth. Cell Rep 38, 110290.
Chartschenko, E., Hugenroth, M., Akhtar, I., Droste, A., Kolkhof, P., Bohnert, M., Beller, M. (2021). CG32803 is the fly homolog of LDAF1 and influences lipid storage in vivo. Insect Biochem Mol Biol. 133, 103512.
Bohnert M. (2020). Tether me, tether me not – Dynamic organelle contact sites in metabolic rewiring. Dev Cell 54, 212-225. (Review)
Castro, I.G., Eisenberg-Bord, M., Persiani, E., Rochford, J.J., Schuldiner, M., Bohnert, M. (2019). Promethin is a conserved seipin partner protein. Cells 8, E268.
Eisenberg-Bord, M., Mari, M., Weill, U., Rosenfeld-Gur, E., Moldavski, O., Castro, I.G., Soni, K.G., Harpaz, N., Levine, T.P., Futerman, A.H., Reggiori, F., Bankaitis, V.A., Schuldiner, M., Bohnert, M. (2018). Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J Cell Biol 217, 269-282.
Rampelt, H., Wollweber, F., Gerke, C., Bohnert, M., Pfanner, N., van der Laan, M. (2018). Assembly of the mitochondrial cristae organizer Mic10 is regulated by Mic26-Mic27 antagonism and cardiolipin. J Mol Biol 430, 1883-1890.
González Montoro, A., Auffahrt, K., Hönscher, C., Bohnert, M., Becker, T., Warscheid, B., Reggiori, F., van der Laan, M., Fröhlich, F., Ungermann, C. (2018). Vps39 interacts with Tom40 to establish one of two functionally distinct vacuole-mitochondria contact sites. Dev Cell 45, 621-636.
Zerbes, R.M., Höß, P., Pfanner, N., van der Laan, M., Bohnert, M. (2016). Distinct roles of Mic12 and Mic27 in the mitochondrial contact site and cristae organizing system. J. Mol. Biol. 428, 1485-1492.
Bohnert, M., Zerbes, R.M., Davies, K.M., Mühleip, A.W., Rampelt, H., Horvath, S.E., Boenke, T., Kram, A., Perschil, I., Veenhuis, M., Kühlbrandt, W., van der Klei, I.J., Pfanner, N., van der Laan, M. (2015). Central Role of Mic10 in the Mitochondrial Contact Site and Cristae Organizing System. Cell Metab 21, 747-755.