Project P13 | Jacob Piehler
Regulation of cytokine receptor signaling plasticity by the subcellular membrane context
We focus on class I/II cytokine receptors with important functions in hematopoiesis and immunity to understand how receptor assembly and effector recruitment is regulated by membrane properties and subcellular localization.
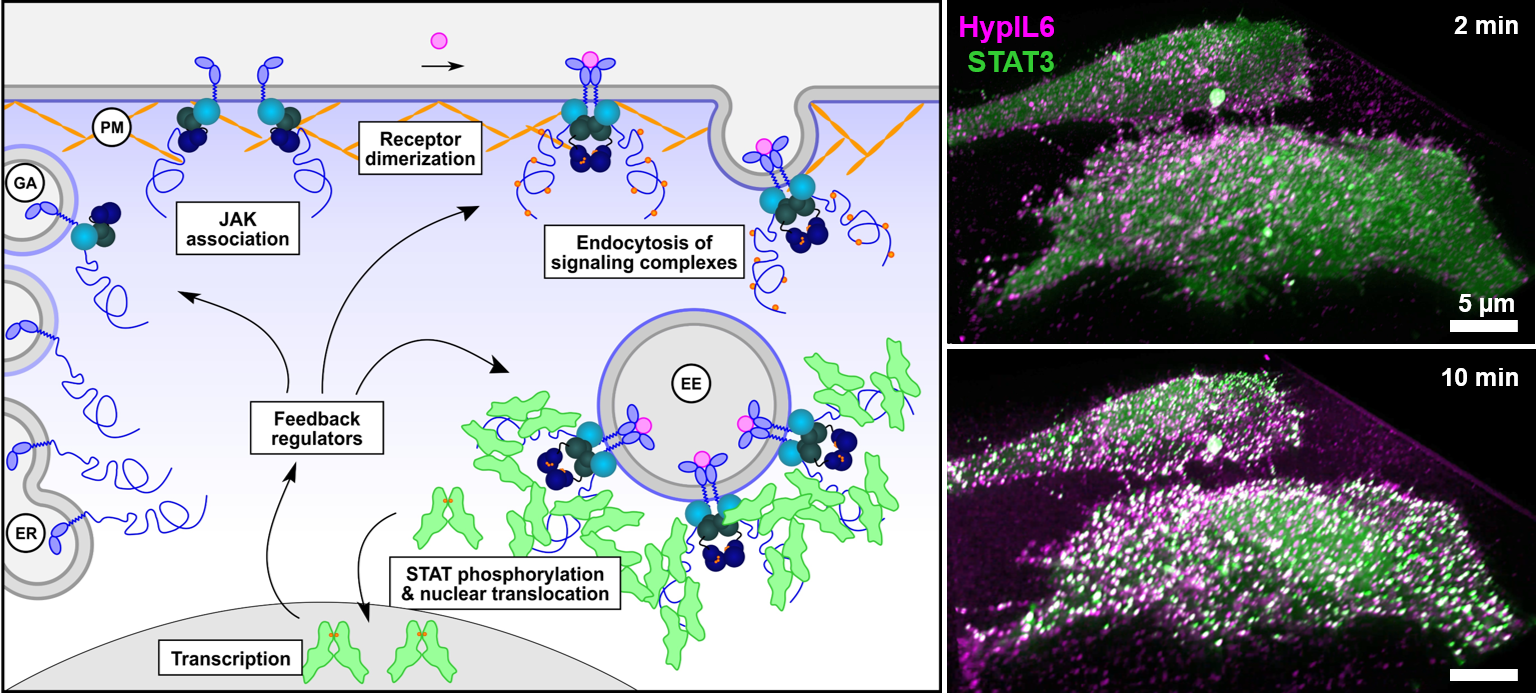
Assembly and activation of cytokine receptors in the context of subcellular compartments. P13 will analyze, how non-covalent association of Janus-familiy kinases (JAKs), receptor dimerization and the recruitment of signal transducer and activator of transcription (STAT) proteins as key downstream effectors are regulated by differential membranes properties within the endomembrane system. © Jacob Piehler
Project Summary
Prof. Dr. Jacob Piehler
Osnabrück University
School of Biology/Chemistry
Research Group Biophysics

Class I/II cytokines are key regulators of hematopoiesis and immune responses. Their receptors (CRs) activate downstream signaling pathways via Janus family tyrosine kinases (JAKs), which are non-covalently associated via membrane-proximal motifs of the cytoplasmic domains of the receptors.
Downstream signaling and cellular responses are mostly governed by tyrosine phosphorylation of different signal transducer and activator of transcription (STAT) proteins, but other signaling pathways are involved to fine-tune cellular decisions. Interestingly, signaling signatures and cellular responses elicited by individual CRs can be strongly modulated by different ligands and cellular settings.
We have demonstrated that the spatiotemporal regulation of receptor activity is a key determinant of such functional plasticity of CR signaling. By quantitative single molecule imaging techniques, we have identified key mechanistic principles of CR assembly and activation in the plasma membrane.
Our previous studies have identified potential regulatory cues related to subcellular membrane properties and we hypothesize that these have important implications in defining context-specific signature of CR signaling. Based on these insights, this project comes to explore how the plasticity of CR signaling is encoded by the changing subcellular membrane environment encountered by signaling complexes during trafficking.
Project-related publications
Sotolongo Bellón, J., Birkholz, O., Richter, C.P., Eull, F., Kenneweg, H., Wilmes, S., Rothbauer, U., You, C., Walter, M.R., Kurre, R., Piehler, J. (2022). Four-color single-molecule imaging with engineered tags resolves the molecular architecture of signaling complexes in the plasma membrane. Cell Reports Methods 2, 100165.
Wilmes, S., Jeffrey, P.A., Martinez-Fabregas, J., Hafer, M., Fyfe, P.K., Pohler, E., Gaggero, S., Lopez-Garcia, M., Lythe, G., Taylor, C., Guerrier, T., Launay, D., Mitra, S., Piehler, J., Molina-Paris, C., Moraga, I. (2021). Competitive binding of STATs to receptor phospho-Tyr motifs accounts for altered cytokine responses. eLife 10, e66014.
Glassman, C.R., Su, L., Majri-Morrison, S.S., Winkelmann, H., Mo, F., Li, P., Perez-Cruz, M., Ho, P.P., Koliesnik, I., Nagy, N., Hnizdilova, T., Picton, L.K., Kovar, M., Bollyky, P., Steinman, L., Meyer, E., Piehler, J., Leonard, W.J., Garcia, K.C. (2021). Calibration of cell-intrinsic interleukin-2 response thresholds guides design of a regulatory T cell biased agonist. eLife 10, e65777.
Gorby, C., Sotolongo Bellon, J., Wilmes, S., Warda, W., Pohler, E., Fyfe, P.K., Cozzani, A., Ferrand, C., Walter, M.R., Mitra, S., Piehler, J., Moraga, I. (2020). Engineered IL-10 variants elicit potent immunomodulatory effects at low ligand doses. Sci Signal 13.
Wilmes, S., Hafer, M., Vuorio, J., Tucker, J.A., Winkelmann, H., Lochte, S., Stanly, T.A., Pulgar Prieto, K.D., Poojari, C., Sharma, V., Richter, C.P., Kurre, R., Hubbard, S.R., Garcia, K.C., Moraga, I., Vattulainen, I., Hitchcock, I.S., Piehler, J. (2020). Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science 367, 643-652.
Martinez-Fabregas, J., Wilmes, S., Wang, L., Hafer, M., Pohler, E., Lokau, J., Garbers, C., Cozzani, A., Fyfe, P.K., Piehler, J., Kazemian, M., Mitra, S., Moraga, I. (2019). Kinetics of cytokine receptor trafficking determine signaling and functional selectivity. eLife 8, e49314.
Mendoza, J.L., Escalante, N.K., Jude, K.M., Sotolongo Bellon, J., Su, L., Horton, T.M., Tsutsumi, N., Berardinelli, S.J., Haltiwanger, R.S., Piehler, J., Engleman, E.G., Garcia, K.C. (2019). Structure of the IFNgamma receptor complex guides design of biased agonists. Nature 567, 56-60.
Richter D, Moraga I, Winkelmann H, Birkholz O, Wilmes S, Schulte M, Kraich M, Kenneweg H, Beutel O, Selenschik P, Paterok D, Gavutis M, Schmidt T, Garcia KC, Muller TD, Piehler J (2017) Ligand-induced type II interleukin-4 receptor dimers are sustained by rapid re-association within plasma membrane microcompartments. Nat Commun 8:15976.
Wilmes S, Beutel O, Li Z, Francois-Newton V, Richter CP, Janning D, Kroll C, Hanhart P, Hotte K, You C, Uze G, Pellegrini S, Piehler J (2015) Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J Cell Biol 209(4):579-593.